Acta medica Lituanica ISSN 1392-0138 eISSN 2029-4174
2022. Online ahead of print DOI: https://doi.org/10.15388/Amed.2021.29.1.7
Analysis of Epigenetic Changes in Vitamin D Pathway Genes in Rheumatoid Arthritis Patients
Eglė Puncevičienė*
ORCID ID: https://orcid.org/0000-0002-6229-4177
Clinic of Rheumatology, Orthopaedics Traumatology and Reconstructive Surgery, Institute of Clinical Medicine of the Faculty of Medicine, Vilnius University, Vilnius, Lithuania;
State Research Institute Centre for Innovative Medicine, Vilnius, Lithuania;
Center of Rheumatology, Vilnius University Hospital Santaros klinikos, Vilnius, Lithuania
Justina Gaiževska
ORCID ID: https://orcid.org/0000-0001-9594-9216
Institute of Biosciences, Life Sciences Center, Vilnius University, Vilnius, Lithuania;
National Cancer Institute, Vilnius, Lithuania
Rasa Sabaliauskaitė
ORCID ID: https://orcid.org/0000-0002-3465-3558
National Cancer Institute, Vilnius, Lithuania
Kristina Šnipaitienė
ORCID ID: https://orcid.org/0000-0001-9766-7108
Institute of Biosciences, Life Sciences Center, Vilnius University, Vilnius, Lithuania;
National Cancer Institute, Vilnius, Lithuania
Lina Vencevičienė
ORCID ID: https://orcid.org/0000-0002-1166-7199
Clinic of Internal Medicine, Family Medicine and Oncology, Faculty of Medicine, Vilnius University, Vilnius, Lithuania;
Center of Family Medicine, Vilnius University Hospital Santaros Klinikos, Vilnius, Lithuania
Dalius Vitkus
ORCID ID: https://orcid.org/0000-0003-0912-4125
Institute of Biomedical Sciences of the Faculty of Medicine, Vilnius University, Vilnius, Lithuania;
Center of Laboratory Medicine, Vilnius University Hospital Santaros klinikos, Vilnius, Lithuania
Sonata Jarmalaitė
ORCID ID: https://orcid.org/0000-0003-3719-0891
Institute of Biosciences, Life Sciences Center, Vilnius University, Vilnius, Lithuania;
National Cancer Institute, Vilnius, Lithuania
Irena Butrimienė
ORCID ID: https://orcid.org/0000-0002-3795-7741
Clinic of Rheumatology, Orthopaedics Traumatology and Reconstructive Surgery, Institute of Clinical Medicine of the Faculty of Medicine, Vilnius University, Vilnius, Lithuania;
State Research Institute Centre for Innovative Medicine, Vilnius, Lithuania;
Center of Rheumatology, Vilnius University Hospital Santaros klinikos, Vilnius, Lithuania
Abstract. Background: Rheumatoid arthritis (RA) is an autoimmune inflammatory disease with complex etiopathogenesis launched by multiple risk factors, including epigenetic alterations. RA is possibly linked to vitamin D that is epigenetically active and may alter DNA methylation of certain genes. Therefore, the study aimed to evaluate the relationship between DNA methylation status of vitamin D signaling pathway genes (VDR, CYP24A1, CYP2R1), vitamin D level and associations with RA.
Materials and Methods: Totally 76 participants (35 RA patients and 41 healthy controls) were enrolled from a case-control vitamin D and VDR gene polymorphisms study regarding age and vitamin D concentration. CpG islands in promoter regions of the VDR, CYP24A1, CYP2R1 genes were chosen for DNA methylation analysis by means of pyrosequencing. Chemiluminescent microplate immunoassay was used to assess 25(OH)D serum levels. RA clinical data, i.e. the disease activity score C-reactive protein 28 (DAS28 – CRP) as well as patient-reported outcome questionnaires were recorded.
Results: The study showed similar methylation pattern in the promoter regions of vitamin D pathway genes in RA and control group with p>0.05 (VDR gene 2.39% vs. 2.48%, CYP24A1 gene 16.02% vs. 15.17% and CYP2R1 2.53% vs. 2.41%). CYP24A1 methylation intensity was significantly higher in compare to methylation intensity of VDR and CYP2R1 genes in both groups (p<0.0001). A tendency of higher vitamin D concentration in cases having methylated VDR (57.57±28.93 vs. 47.40±29.88 nmol/l), CYP24A1 (53.23±26.22 vs. 48.23±34.41 nmol/l) and CYP2R1 (60.41±30.73 vs. 44.54±27.63 nmol/l) genes and a positive correlation between VDR, CYP2R1 methylation intensity and vitamin D level in RA affected participants was revealed (p>0.05). A significantly higher CYP24A1 methylation intensity (p=0.0104) was detected in blood cells of vitamin D deficient (<50 nmol/l) RA patients vs. vitamin D deficient controls.
Conclusions: Our data suggests some indirect associations between DNA methylation status of vitamin D pathway genes and vitamin D level in RA.
Keywords: DNA methylation, Rheumatoid arthritis, Vitamin D
Vitamino D atsako kelio genų epigenetinių pokyčių analizė sergantiems reumatoidiniu artritu pacientams
Santrauka. Įvadas. Reumatoidinis artritas (RA) yra sudėtingos etiopatogenezės autoimuninė uždegiminė liga, nulemta daugelio rizikos veiksnių, kuriems priskiriamos ir epigenetinės modifikacijos. Manoma, kad RA yra susijęs su vitaminu D, kuris yra epigenetiškai aktyvus ir gali daryti įtaką tam tikrų genų DNR metilinimui. Šiuo tyrimu siekta įvertinti vitamino D atsako kelio genų (VDR, CYP24A1, CYP2R1) DNR metilinimo sąsajas su vitamino D kiekiu bei RA.
Medžiaga ir metodai. Į tyrimą iš viso įtraukti 76 tiriamieji (35 RA pacientai ir 41 kontrolinės grupės asmuo), pagal amžių ir vitamino D koncentraciją kraujo serume atrinkti iš VDR geno polimorfizmų atvejo kontrolės tyrimo. VDR, CYP24A1, CYP2R1 genų promotorių CpG salų metilinimo analizė atlikta pirosekoskaitos metodu. Vitamino D koncentracijos kraujo serume tyrimas atliktas naudojant chemiliuminescencinę imuninę analizę. Surinkti RA klinikiniai duomenys: ligos aktyvumo indeksas vertinant 28 sąnarius (angl. Disease activity score, DAS28) ir būklės įsivertinimo klausimynų duomenys.
Rezultatai. Tyrimo metu buvo rastas panašus RA ir kontrolinės grupės tiriamųjų vitamino D apykaitos genų metilinimo lygis (VDR geno 2,39 % vs 2,48 %, CYP24A1 geno 16,02 % vs 15,17 % ir CYP2R1 geno 2,53 % vs 2,41 %). CYP24A1 metilinimo intensyvumas buvo statistiškai reikšmingai didesnis, palyginti su VDR ir CYP2R1 genų metilinimo intensyvumu abiejose grupėse (p<0,0001).Tendencingai didesnė vitamino D koncentracija nustatyta RA tiriamiesiems, kurių genai buvo metilinti palyginus su nemetilintais atvejais: VDR geno (57,57 ± 28,93 vs 47,40 ± 29,88 nmol/l), CYP24A1 (53,23 ± 26,22 vs 48,23 ± 34,41 nmol/l), CYP2R1 geno (60,41 ± 30,73 vs 44,54 ± 27,63 nmol/l); VDR ir CYP2R1 metilinimo intensyvumas teigiamai koreliavo su vitamino D koncentracija RA tiriamųjų grupėje (p>0,05). Statistiškai reikšmingai didesnis CYP24A1 metilinimo intensyvumas nustatytas periferinio kraujo ląstelėse tų RA pacientų, kuriems rastas vitamino D trūkumas (<50 nmol/l), palyginti su vitamino D stokojančiais sveikais asmenimis (p=0,0104).
Išvados. Remiantis gautais duomenimis galima daryti prielaidą, kad vitamino D apykaitos genų metilinimo būklė yra netiesiogiai susijusi su vitamino D kiekiu sergant RA.
Raktažodžiai: DNR metilinimas, reumatoidinis artritas, vitaminas D
__________
* Corresponding author: Puncevičienė Eglė, Address: Center of Rheumatology, Vilnius University Hospital Santaros klinikos, Santariškių str. 2,
LT-08661 Vilnius, Lithuania; E-mail: epunceviciene@gmail.com; Telephone number: +37061822628; Fax number: +37052365302
Received: 12/09/2021. Revised: 01/12/2021. Accepted: 15/12/2021
Copyright © 2022 Eglė Puncevičienė, Justina Gaiževska, Rasa Sabaliauskaitė, Kristina Šnipaitienė, Lina Vencevičienė, Dalius Vitkus, Sonata Jarmalaitė, Irena Butrimienė. Published by Vilnius University Press.This is an Open Access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Introduction
Rheumatoid arthritis (RA) is one of the most common autoimmune inflammatory arthritis with a complex pathogenesis mainly related to the breakdown of immune tolerance, autoantigen presentation with antigen specific T and B cells activation and aberrant production of inflammatory cytokines [1]. The disease manifests as synovial joint inflammation marked by infiltration of immune cells and damage to the extracellular matrix, leading to pain, disability, joint destruction and function loss [2]. It is known that RA development is associated with multiple risk factors that combine interaction between genetic predisposition, environmental risk and epigenetic changes [3]. However, the etiology and pathogenetic mechanisms of disease remain to be not fully unravelled thus demanding new researches, especially in the field of epigenetics.
Recently, epigenetic alterations were named as potential key players in pathogenesis of RA leading to heritable changes in gene activity and expression, without causing alterations in primary deoxyribonucleic acid (DNA) sequence thus regulating various cells’ behaviour [2, 4]. Changes of methylation pattern in peripheral blood T and B cells have been demonstrated in previous reports and suggest that epigenetic variation may mediate pathogenic activity of immune cells in RA [5-7]. These mechanisms lead to increased production of pro-inflammatory cytokines (such as interleukin (IL)-1β, IL-6, tumor necrosis factor (TNF)-α, etc.) and mediate development of chronic inflammation hereafter strengthening the hypothesis of altered DNA methylation signature in RA [3].
Furthermore, the environmental stress can also be reflected in genome as aberrant epigenetic marks, contributing to gene regulation and RA etiopathogenesis mechanisms [2]. Vitamin D is one of the potential environmental factors participating in RA course through the regulation and differentiation of the immune cells, performing its’ extra-skeletal anti-inflammatory, immuno-modulatory role [8-10]. The metabolism of vitamin D consists of multiple conversions mediated by cytochrome P450 (CYP) enzymes. The enzyme 25-hydroxylase (CYP2R1) converts cholecalciferol to 25-hydroxycholecalciferol (25(OH)D3), 1α-hydroxylase (CYP27B1) converts to active form 1.25-dihydroxyvitamin D (calcitriol) whilst 24-hydroxylase (CYP24A1) inactivates and decreases the calcitriol level either decreases the substrate available for its production [11]. The other important vitamin D signaling system player is the vitamin D receptor (VDR) which is expressed in multiple immune cells. It is also the key mediator in active vitamin D regulated gene expression [12]. Calcitriol interacts with the epigenome on multiple levels, e.g., binds the VDR and by the heterodimerized form further binds vitamin D response elements which lie in the promoter regions of vitamin D responsive genes and in this manner upregulates or suppresses DNA transcription [11, 13]. On the other hand, the above mentioned “vitamin D tool” genes coding VDR, CYP2R1, CYP27A1 and CYP24A1 tend themselves to be epigenetically regulated [13]. Promoter regions of these genes span the Cytosine-phosphate-Guanine (CpG) dinucleotide islands and thus can be silenced by DNA methylation [13, 14]. It is known that vitamin D deficiency is prevalent in RA patients, is associated with higher disease activity and the level appears to be significantly lower compared to healthy controls [15, 16]. Hypothesis of higher risk to develop RA in vitamin D deficient subjects has also been raised [17]. Therefore, since the attention to vitamin D role in RA is increasing and 25(OH)D signaling pathway has been poorly investigated in RA subjects, there is a need to analyze the methylation pattern of vitamin D metabolism genes in this group of patients.
The present study evaluated vitamin D pathway genes (VDR, CYP2R1, CYP24A1) methylation level in peripheral blood mononuclear cells of RA subjects and healthy controls in order to identify novel vitamin D-RA associated DNA methylation sites, and the relationship between selected genes methylation status with vitamin D level and RA clinical parameters.
Materials and methods
Study cohorts
Totally 76 participants (35 RA patients and 41 healthy control subject) were included from a case-control vitamin D and VDR gene polymorphisms study regarding age and vitamin D concentration [18]. All participants were enrolled at Vilnius University Hospital Santaros Klinikos (VUHSK) Rheumatology Centre after informed consent was obtained according to the permission of Vilnius Regional Biomedical Research Ethics Committee (Approval No. 158200-18/5-1037-533). RA diagnosis was established by ACR/EULAR 2010 rheumatoid arthritis classification criteria or by 1987 ACR classification criteria if diagnosed before year 2010 [19, 20]. Researchers collected sociodemographic (sex, age, body mass index, smoking status, etc.) and clinical data from all RA patients (disease duration, present treatment, disease activity score 28 (DAS28), C-reactive protein (CRP, mg/l), etc.). The DAS28 CRP was measured by a researcher rheumatologist and categorized accordingly as high (≥5.1), moderate (>3.2 to 5.1), low disease activity (2.6 to ≤3.2) or RA remission (<2.6) [21]. All eligible control group subjects were invited to participate at VUHSK Family Medicine Centre by a family doctor and further referred to Rheumatology Centre for informed consent form signing, data and blood samples collection.
All recruited subjects were adults over 18 years old. Subjects who did not meet the inclusion criteria were excluded (diagnosed cancer (<5 years), other autoimmune comorbidities, pregnancy, etc.) from the study. For biochemical tests and epigenetic analysis, blood samples from all study subjects were collected, coded and labeled as required. The research was carried out in accordance with the principles of the Declaration of Helsinki of 1975, revised in 2013.
Cells preparation
Blood samples for DNA analysis from RA and control group were collected using BD Vacutainer® CPTTM Mononuclear Cell Preparation Tube – Sodium Citrate vacutainers (8 mL) (BD Biosciences, Franklin Lakes, NJ, USA). Peripheral blood mononuclear cells (PBMCs) were prepared using manufacturer’s recommendations and standardized procedures: tubes with blood samples were centrifuged (Centrifuge – Heraeus Megafuge 8 Centrifuge, Thermo Scientific (TS) part of Thermo Fisher Scientific (TFS), Wilmington, DE, USA) by stages and sequential cell washing steps adding phosphate-buffered saline (PBS) (Biochrom, Berlin, Germany) were performed as recommended. Samples with prepared PBMCs were labelled and stored at –80°C temperature until DNA methylation analysis.
DNA methylation analysis
All 76 samples selected for DNA methylation analysis were age, sex, and vitamin D matched. DNA isolation was performed using commercial GeneJET Genomic DNA Purification Kit (TFS) by manufacturer’s recommendations. Isolated DNA concentration and purity was measured by 260 / 280 and 260 / 230 ratios using NanoDrop® 2000 spectrophotometer (TS, TFS). Each subject DNA sample was bisulphate converted using EZ DNA Methylation™ Kit (Zymo Research, CA, USA) by manufacturer’s recommendations. Converted DNA samples were labelled and stored at –20°C. CpG islands in promoter regions of VDR, CYP24A1, CYP2R1 genes were chosen for DNA methylation analysis. Primers for selected genes polymerase chain reaction (PCR) and pyrosequencing were designed using Ensembl data base (https://www.ensembl.org/index.html) and PyroMark Assay Design Software (version 2.0.1.15, Qiagen, 2008, Germany). PCR and pyrosequencing primers are listed in Supplementary Material, Table S1. PCR method was used to amplify modified DNA. PCR cycling conditions are shown in Supplementary Material, Table S2. The success of DNA amplification was assessed by 3% agarose gel electrophoresis. Methylation was evaluated in 10 VDR gene CpG positions, 7 CYP24A1 and 6 CYP2R1 CpG positions by means of pyrosequencing of PCR products and performed using PyroMark Q24 platform (Qiagen, Berlin, Germany) by manufacturer’s recommendations. Methylation intensity of each CpG site was generated using PyroMark Q24 Software (version 2.0.6, Qiagen, 2009, Germany) and assessed as percentage.
Vitamin D evaluation
Blood samples for 25(OH)D measurement from RA and control group were collected using BD Vacutainer Serum Separator Tubes (5 mL) (BD Biosciences, New Jersey, USA) and evaluated at VUHSK Centre of Laboratory medicine using chemiluminescent microplate immunoassay (Architect ci8200, Abbott Laboratories, IL, USA), with the ability to detect 25(OH)D3 from 98.6% to 101.1% and 25(OH)D2 from 80.5% to 84.4%. Vitamin D concentration was classified as normal (≥75 nmol/l), insufficient (≥50–75 nmol/l) or deficient (<50 nmol/l) according to the Endocrine Society clinical practice guideline [22]. To avoid sources of bias blood samples collection was performed from late October until mid-May.
Data statistical analysis
Descriptive statistics for demographic, clinical and biochemical variables was applied. Baseline methylation levels of VDR, CYP24A1 and CYP2R1genes were calculated. After that, qualitative and quantitative methylation analysis of listed genes’ promoters CpG sites was performed. After Shapiro–Wilk normality test was applied, a parametric paired t-test or non parametric Mann–Whitney rank-sum test was performed for quantitative DNA methylation level (methylation intensity) analysis. To assess associations for qualitative DNA methylation level (methylation frequency) the Fisher’s exact or Chi-square test was used. Spearman rank-order correlation analysis was applied for demographic, clinical, biochemical parameters and VDR, CYP24A1, CYP2R1 genes promoter’smethylation level.
Statistical analysis and data visualization was performed with R (Version 1.1.383, RStudio Inc, Boston, MA, USA), Microsoft Office Excel 2016 (Microsoft Corporation, Redmond, WA, USA) GraphPad Prism (version 8.0.1, GraphPad Software, San Diego, CA, USA) and ClustVis web tool [23]. Values were considered to be statistically significant at p<0.05.
Results
Main study population characteristics
From a case-control vitamin D and VDR gene polymorphisms study [18] a group of 35 RA patients and 41 healthy controls was selected for further DNA methylation analysis of vitamin D metabolism pathway genes. Average age of RA subjects at enrolment into the study was 51.63±10.45 years, mean disease duration 12.40±8.22 years. For comparison analysis, age, sex and vitamin D matched healthy controls with mean age of 50.78±11.85 years were enrolled. In order to precisely assess epigenetic differences in both groups, measured serum concentration of 25(OH)D in RA patients did not differ significantly in comparison with controls (50.89±29.54 vs. 56.05±28.67, p>0.05). The detailed descriptive analysis of anthropometric, sociodemographic and clinical data of study participants is presented in Table 1.
Table 1. Characteristics of study participants
|
Characteristics |
RA, N=35 |
Healthy Controls, N=41 |
p value |
|
Sex (N, %) |
|||
|
Female Male |
34 (97.1) 1 (2.9) |
40 (97.5) 1 (2.5) |
0.91 |
|
Age (years) |
51.63±10.45 |
50.78±11.85 |
0.74 |
|
BMI (kg/m2) |
25.60±4.39 |
26.22±5.51 |
0.99 |
|
Smoking status (N, %) |
|||
|
Current smokers Former smokers Never smokers |
3 (8.6) 4 (11.4) 28 (80) |
2 (4.9) 8 (19.5) 31 (75.6) |
0.54 |
|
Disease activity (DAS28 CRP score) |
4.20±1.45 |
||
|
High (N, %) Moderate (N, %) Low (N, %) Remission (N, %) |
7 (20) 20 (57.2) 4 (11.4) 4 (11.4) |
N.A. |
N.A. |
|
Disease duration (years) |
12.40±8.22 |
N.A. |
N.A. |
|
Vitamin D (nmol/l) |
50.89±29.54 |
56.05±28.67 |
|
|
Deficiency (<50 nmol/l) (N, %) Insufficiency (≥50 -75 nmol/l) (N, %) Normal (≥75 nmol/l)(N, %) |
16 (45.7) 10 (28.6) 9 (25.7) |
17 (41.5) 11 (26.8) 13 (31.7) |
0.2745 |
|
CRP (mg/l) |
6.32±7.12 |
N.A. |
N.A. |
BMI: body mass index; CRP: C reactive protein; DAS28: disease activity score 28; RA: rheumatoid arthritis; N.A.; not applicable. Data represent mean ± standard deviation or N, %.
DNA methylation analysis in peripheral blood mononuclear cells in RA patients and controls
In order to assess the methylation differences between the cases and controls, as well as its associations with RA clinical parameters and vitamin D level, DNA methylation level analysis of VDR, CYP24A1, CYP2R1 promoters was performed by means of pyrosequencing.
VDR, CYP24A1, CYP2R1 methylation pattern
The mean methylation intensity of analyzed genes in RA vs. control group was as follows: VDR gene 2.39% vs. 2.48%, CYP24A1 gene 16.02% vs. 15.17% and CYP2R1 2.53% vs.2.41%. According to the average methylation intensity of each gene promoter regions, the threshold of methylation level was specified, and methylated vs. unmethylated samples detected to access the methylation frequency of all analyzed CpG positions (Figure 1A and 1B). Although the differences between the cases and controls were statistically insignificant (p>0.05), CYP24A1 methylation intensity was significantly higher compared to methylation intensity of VDR and CYP2R1 genes promoters in both groups (p<0.0001) (Figure 1C and 1D).
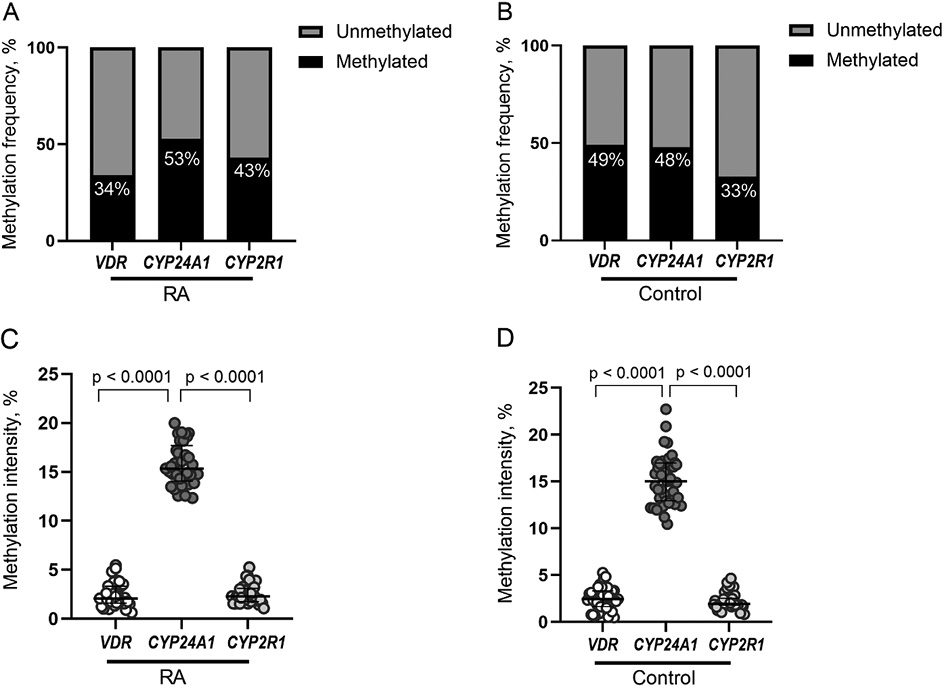
Figure 1. VDR, CYP24A1 and CYP2R1 promoter methylation frequencies (percentages) in RA (A) and healthy controls (B). Methylation intensity comparison of VDR, CYP24A1 and CYP2R1 genes promoters in RA patients (C) and healthy controls (D). RA: rheumatoid arthritis.
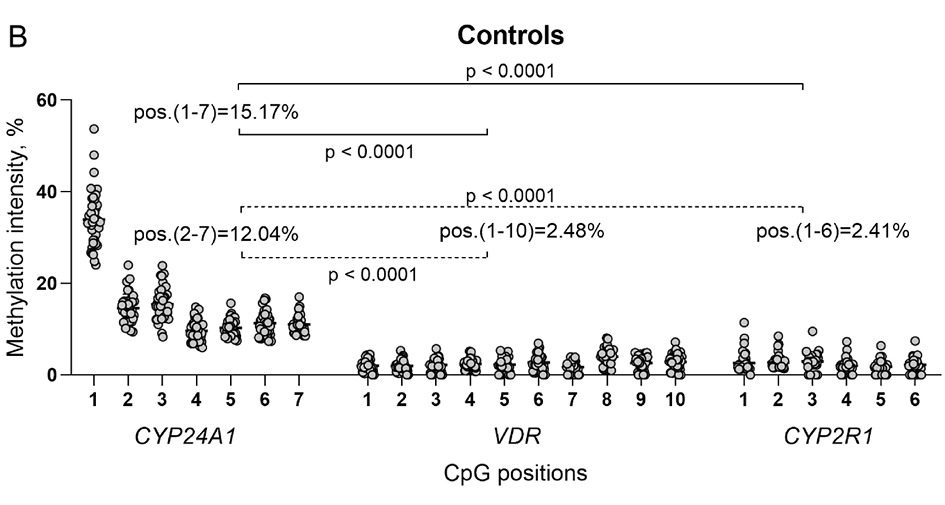
The study also applied hierarchical clustering heatmap method to evaluate the methylation intensity of selected individual CpG positions located in promoter regions of VDR, CYP24A1 and CYP2R1 genes. No significant differences between RA and control group were detected (p>0.05). However, the comparison of separate CpG positions in RA revealed several significant differences. Significantly higher methylation intensity of VDR 8 CpG position compared to 1–4 and 7th positions was detected (1 vs. 8 p<0.0001; 2 vs. 8 p<0.0001; 3 vs. 8 p=0.0060; 4 vs. 8 p=0.0014; 7 vs. 8 p=0.0032). Also, the 1st CpG position of CYP24A1 gene showed higher methylation intensity level in comparison with 2–7 positions (p < 0.0001) and 3rd position in comparison with 4–7 CpG positions (3 vs. 4 p<0.0001; 3 vs. 5 p=0.0005; 3 vs. 6 p=0.0079; 3 vs. 7 p=0.0020). Furthermore, 3rd CpG position of the CYP2R1gene was more intensively methylated than 1–5 positions (1 vs. 3 p=0.0019; 2 vs. 3 p=0.0286; 3 vs. 4 p=0.0003; 3 vs. 5 p=0,0035). Similar results were obtained from the control group analysis as well (Figure 2A, 2B and 2C). The graph with a more detailed information on mean methylation values of each gene CpG position and differences between the genes promoter regions in RA and controls is presented the Supplementary Material Figure S1 A and B.
Hereafter, associations between selected genes promoters’ methylation pattern and demographic features in RA group have been analyzed. CYP24A1 methylation level positively correlated with age in RA patients (r=0.519, p=0.0017). Also, after comparing methylation frequencies, CYP24A1 gene methylation was found to be significantly more frequent in older RA vs. younger participants (p<0.01). However, similar methylation changes were also seen in control group, possibly indicating universal age-related changes (p=0.01). No other significant associations between demographic parameters (i.e. sex, smoking status, etc.) and methylation pattern were discovered.
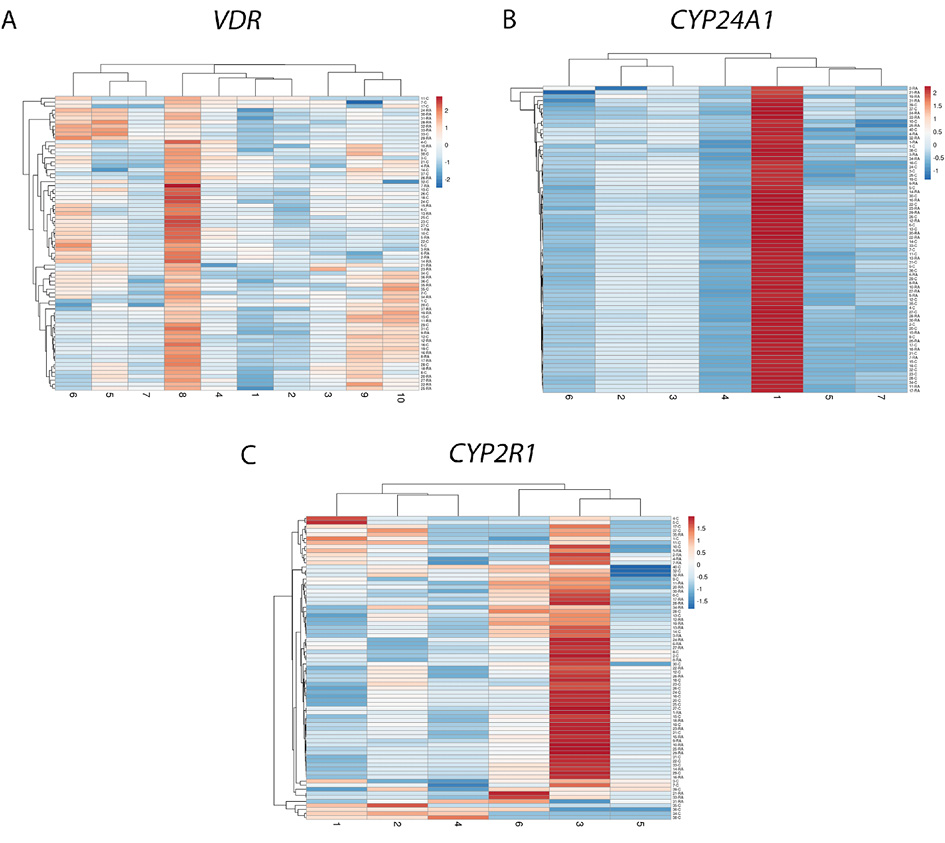
Figure 2. CpG methylation intensity comparison of VDR (A), CYP24A1 (B), CYP2R1 (C) promoters’ separate positions (X-axis) in RA and control group samples (Y-axis). Hierarchical clustering heatmap obtained using ClustVis web tool [23]. RA: rheumatoid arthritis. The blue-to-red color scale corresponds to the lowest and highest methylation intensity values.
DNA methylation and RA clinical parameters
The association of DNA methylation and RA disease clinical parameters was evaluated in the study. A tendency of higher RA DAS28 CRP disease activity score in CYP24A1 methylated vs. unmethylated gene promoter RA cases (3.71±0.891 vs. 4.60±0.48, p=0.0774) was found (Figure 3A). However, the study found no significant associations between selected genes DNA methylation intensity and DAS28 CRP score in RA group. Nevertheless, a higher RAID score was significantly associated with VDR gene promoter methylation status (4.58±1.64 vs. 6.22±0.66, p=0.018) (Figure 3B). Also, VDR, as well as CYP2R1, methylated vs. unmethylated promoter cases had a higher HAQ score (0.82±0.20 vs. 1.05±0.23 and 0.85±0.19 vs. 0.97±0.12, respectively), however it was not statistically significant (Figure 3C). Other RA clinical variables, such as CRP, did not show any significant differences in respect of gene’s methylation level.
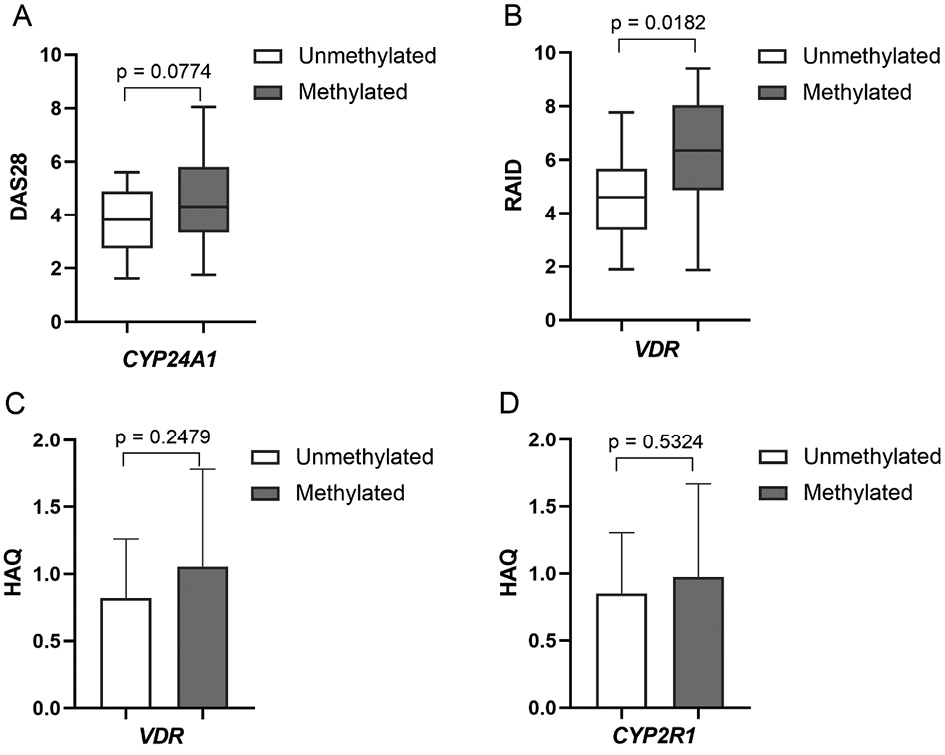
Figure 3. Comparison of cases with methylated vs. unmethylated CYP24A1, VDR and CYP2R1 genes promoters and RA clinical parameters: DAS28 score (A), RAID score (B) and HAQ score (C). DAS28: disease activity score; HAQ: health assessment questionnaire; RA: rheumatoid arthritis; RAID: rheumatoid arthritis impact of disease.
DNA methylation and vitamin D level
The study data showed higher vitamin D concentration in RA cases with methylated vs. unmethylated VDR (57.57±28.93 vs. 47.40±29.88 nmol/l), CYP24A1 (53.23±26.22 vs. 48.23±34.41 nmol/l) and CYP2R1(60.41±30.73 vs. 44.54±27.63 nmol/l) gene promoters, indicating that DNA methylation pattern of afore mentioned genes tend to be associated with higher vitamin D level. The differences were not statistically significant (Figure 4A), however, methylation intensity analysis of separate CpG positions revealed a significant positive correlation of VDR 8th position and vitamin D level (r=0.3485, p=0.0345) in RA (Figure 4B). Furthermore, methylation intensity of VDR and CYP2R1 genes promoters positively correlated with vitamin D concentration in RA patients, though not significantly (p>0.05). Vitamin D deficient (<50 nmol/l) RA subjects revealed a significantly higher CYP24A1 methylation intensity vs. vitamin D deficient controls (p=0.0104), thus indicating the disturbed vitamin D metabolism in RA (Figure 4C). Additionally, a higher CYP2R1 methylation intensity was also revealed in vitamin D deficient, as well as normal vitamin D level RA vs. control group, respectively, however not significant (Figure 4D). The methylation level of selected genes promoters did not differ significantly according to vitamin D status (normal range (≥75 nmol/l) vs. insufficiency (≥50–75 nmol/l) or deficiency (<50 nmol/l) and vice versa) in RA group. The use of vitamin D supplementation also did not show any statistically significant differences between the groups.
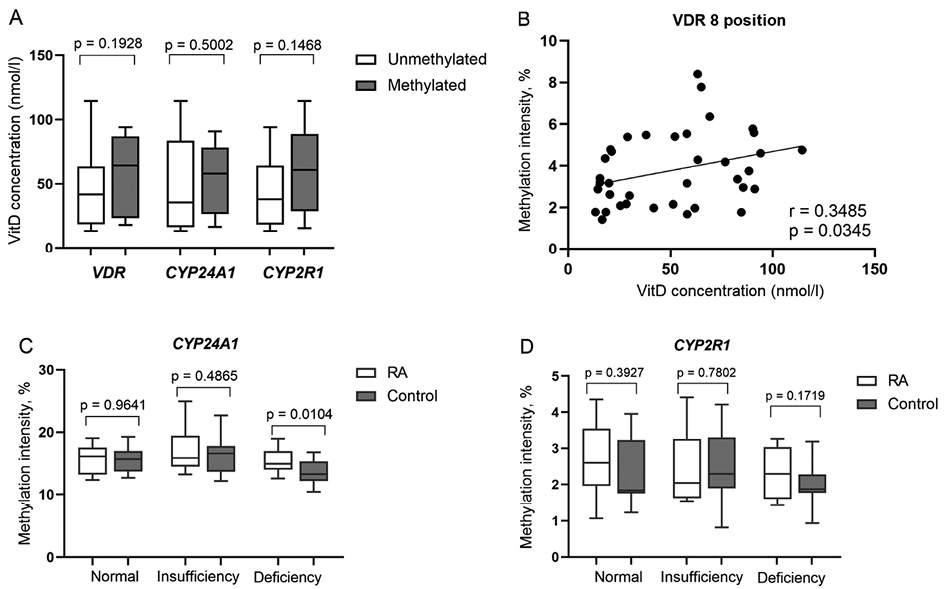
Figure 4. Vitamin D level (nmol/l) comparison by VDR, CYP24A1 and CYP2R1 genes promoter methylation frequency in RA subjects (A); VDR 8th CpG position methylation intensity (percentages) correlation with vitamin D level (nmol/l) (B); Comparison of methylation intensity of CYP24A1 (C) and CYP2R1 (D) promoters according to vitamin D level (nmol/l) in RA subjects and healthy controls. RA: rheumatoid arthritis; VitD: vitamin D
Discussion
The accumulating evidence indicates that epigenetic changes are stated as possible contributors in complex RA mechanisms, biomarkers for diagnosis or even therapeutic targets [3, 24]. Vitamin D acting as immunomodulator in various cell behavior mechanisms might be responsible for epigenetic modifications and consequently contribute to RA pathogenesis [12, 13, 25]. However, none of previously reported studies have analyzed DNA methylation pattern of vitamin D pathway genes in RA patients to date. The present study aimed to evaluate methylation level of VDR, CYP24A1 and CYP2R1 genes promoter regions in peripheral blood cells of RA and healthy controls in order to assess the differences between the groups, possible associations of epigenetic changes with RA clinical parameters and vitamin D level.
The study showed similar methylation pattern in the promoter regions of vitamin D pathway genes in RA and control group. Additionally, an overall low DNA methylation level was observed, except for CYP24A1, which revealed a significantly higher mean methylation intensity of approximately 15 to 17 percent in both groups. Similarly to our results, a study analysing VDR methylation in Behcet’s disease affected subjects and healthy controls reported no significant differences between the groups [26]. Our data is also consistent with the study conducted by Beckett et al. that showed low average methylation level of analyzed CYP2R1, CYP24A1 and VDR genes [27]. A higher methylation level of CYP24A1 gene promoter revealed in our study may be related to high prevalence of vitamin D deficiency in both study groups, RA and control, since CYP24A1 enzyme is responsible for degradation of calcitriol – an active form of vitamin D. On the other hand, low CYP2R1 and VDR methylation intensity allows normal gene expression and indirectly indicates active vitamin D metabolism process.
Confirming the hypothesis, our study results showed a tendency of higher vitamin D concentration in RA patients with methylated VDR, CYP24A1, CYP2R1 promoters and a positive correlation between VDR, CYP2R1 methylation intensity and vitamin D level in RA. In accordance to our findings, Zhu H. and colleagues revealed that participants with severe vitamin D deficiency (25(OH)D ≤25 nmol/l) showed a pattern of reduced methylation of CYP2R1, CYP24A1 and DHCR7 genes in comparison with controls (25(OH)D >75 nmol/L) [28]. The study conducted by Beckett et al. also demonstrated a positive plasma 25(OH)D level correlation with VDR gene methylation level, meanwhile CYP2R1 and CYP24A1 methylation, in contrast to our results, negatively correlated with vitamin D concentration [27]. Also, in contrary to our results, a vitamin D (1100 IU/day) and calcium intervention trial identified that the average methylation of CYP2R1 and CYP24A1 genes was negatively associated with 12-month increase in serum 25(OH)D. Interestingly, non responders to vitamin D supplements at baseline had significantly higher CYP2R1 and CYP24A1 methylation levels compared to that in the responders thus predicting vitamin D response [29]. Regarding the results of present study and previous findings, associations demonstrated between 25(OH)D level and CYP24A1, CYP2R1, VDR promoters’ methylation support the relationship between vitamin D and the epigenome.
A more detailed analysis regarding categorized vitamin D level showed some significant findings between the groups. A noteworthy revelation of the present study is the observed higher methylation level of CYP24A1 and CYP2R1 genes promoters in vitamin D deficient (<50 nmol/l) RA participants vs. vitamin D deficient control group. Furthermore, RA patients with normal vitamin D concentration (≥75 nmol/l) presented higher CYP2R1 methylation level vs. equivalent vitamin D level control subgroup. Although, the DNA methylation profile in RA tends to be globally hypomethylated rather than hypermethylated in PBMCs and RA fibroblast-like synoviocytes [30, 31], DNA methylation changes that are demonstrated in our study could prompt considerable reasons of higher vitamin D deficiency prevalence in RA subjects. This could be explained by hypoproduction of vitamin D metabolism enzymes induced by gene silencing, as mentioned before.
Despite the homogeneously formed RA and control groups regarding age and vitamin D level, the study showed no significant differences of vitamin D pathway genes between the groups. Notably, this suggests that the methylation pattern of vitamin D pathway genes is contingently associated with lower vitamin D level in RA. Also, it is known that the presence of 25(OH)D metabolism enzymes, except for VDR, is more relevant in other cells than PBMCs [12], thus prompting that the methylation level of selected genes may differ in various tissues. Furthermore, one of the potential drawbacks of our study was the limited sample size and overall high prevalence of suboptimal vitamin D level in both groups. The above mentioned factors could have affected the significance of analyzed data. Moreover, causal relationships between the variables could not be assessed due to the study design. Despite the careful technical procedures, the methylation status of the promoters was determined by analysis of selected CpGs, which might not completely reflect the methylation level of entire region. Also, other genes and other regulatory mechanisms involved in regulation of vitamin D pathway should be included in further studies of vitamin D role in RA.
In summary, a tendency of higher vitamin D concentration in cases with methylated VDR, CYP24A1, CYP2R1 genes promoters and a positive correlation between VDR, CYP2R1 methylation intensity and vitamin D level in RA affected participants was shown. Vitamin D deficient (<50 nmol/l) RA patients revealed a significantly higher CYP24A1 methylation intensity vs. vitamin D deficient controls. The analyzed data indicate that the methylation pattern of vitamin D metabolism genes is indirectly associated with vitamin D level in RA. Therefore, the necessity of further investigations of vitamin D regulatory mechanisms in RA patients remains to be actual.
Conflicts of Interest
The authors state no conflict of interest.
Funding
The research was funded by the Research Council of Lithuania (LMTLT), agreement No. S-MIP-17-12.
References
- Calabresi E, Petrelli F, Bonifacio AF, Puxeddu I, Alunno A. One year in review 2018: pathogenesis of rheumatoid arthritis. Clin Exp Rheumatol. 2018 Mar-Apr;36(2):175-184.Epub 2018 Apr 18. PMID: 29716677.
- Firestein GS. Pathogenesis of rheumatoid arthritis: the intersection of genetics and epigenetics. Trans Am Clin Climatol Assoc. 2018;129:171-182. PMID: 30166712; PMCID: PMC6116585.
- Ciechomska M, Roszkowski L, Maslinski W. DNA Methylation as a Future Therapeutic and Diagnostic Target in Rheumatoid Arthritis. Cells. 2019 Aug 22;8(9):953. https://doi:10.3390/cells8090953
- Klein K, Gay S. Epigenetics in rheumatoid arthritis. CurrOpinRheumatol. 2015 Jan;27(1):76-82. https://doi:10.1097/BOR.0000000000000128
- Bottini N, Firestein GS. Epigenetics in rheumatoid arthritis: a primer for rheumatologists. CurrRheumatol Rep. 2013 Nov;15(11):372. https://doi:10.1007/s11926-013-0372-9
- Julià A, Absher D, López-Lasanta M, Palau N, Pluma A, Waite Jones L, Glossop JR, Farrell WE, Myers RM, Marsal S. Epigenome-wide association study of rheumatoid arthritis identifies differentially methylated loci in B cells. Hum Mol Genet. 2017 Jul 15;26(14):2803-2811. https://doi:10.1093/hmg/ddx177
- Glossop JR, Emes RD, Nixon NB, Haworth KE, Packham JC, Dawes PT, Fryer AA, Mattey DL, Farrell WE. Genome-wide DNA methylation profiling in rheumatoid arthritis identifies disease-associated methylation changes that are distinct to individual T- and B-lymphocyte populations. Epigenetics. 2014 Sep;9(9):1228-37. https://doi:10.4161/epi.29718
- Jeffery LE, Raza K, Hewison M. Vitamin D in rheumatoid arthritis-towards clinical application. Nat Rev Rheumatol. 2016 Apr;12(4):201-10. https://doi:10.1038/nrrheum.2015.140
- Umar M, Sastry KS, Chouchane AI. Role of Vitamin D Beyond the Skeletal Function: A Review of the Molecular and Clinical Studies. Int J Mol Sci. 2018 May 30;19(6):1618. https://doi:10.3390/ijms19061618
- Orbach H, Zandman-Goddard G, Amital H, Barak V, Szekanecz Z, Szucs G, Danko K, Nagy E, Csepany T, Carvalho JF, Doria A, Shoenfeld Y. Novel biomarkers in autoimmune diseases: prolactin, ferritin, vitamin D, and TPA levels in autoimmune diseases. Ann N Y Acad Sci. 2007 Aug;1109:385-400. https://doi:10.1196/annals.1398.044
- Ong LTC, Booth DR, Parnell GP. Vitamin D and its Effects on DNA Methylation in Development, Aging, and Disease. Mol Nutr Food Res. 2020 Oct 20:e2000437. https://doi:10.1002/mnfr.202000437
- Harrison SR, Li D, Jeffery LE, Raza K, Hewison M. Vitamin D, Autoimmune Disease and Rheumatoid Arthritis. Calcif Tissue Int. 2020 Jan;106(1):58-75. https://doi:10.1007/s00223-019-00577-2
- Fetahu IS, Höbaus J, Kállay E. Vitamin D and the epigenome. Front Physiol. 2014 Apr 29;5:164. https://doi:10.3389/fphys.2014.00164
- Bahrami A, Sadeghnia HR, Tabatabaeizadeh SA, Bahrami-Taghanaki H, Behboodi N, Esmaeili H, Ferns GA, Mobarhan MG, Avan A. Genetic and epigenetic factors influencing vitamin D status. J Cell Physiol. 2018 May;233(5):4033-4043. https://doi:10.1002/jcp.26216
- Meena N, Singh Chawla SP, Garg R, Batta A, Kaur S. Assessment of Vitamin D in Rheumatoid Arthritis and Its Correlation with Disease Activity. J Nat Sci Biol Med. 2018 Jan-Jun;9(1):54-58. https://doi:10.4103/jnsbm.JNSBM_128_17
- Lee YH, Bae SC. Vitamin D level in rheumatoid arthritis and its correlation with the disease activity: a meta-analysis. Clin Exp Rheumatol. 2016 Sep-Oct;34(5):827-833. Epub 2016 Apr 6. PMID: 27049238.
- Song GG, Bae SC, Lee YH. Association between vitamin D intake and the risk of rheumatoid arthritis: a meta-analysis. Clin Rheumatol. 2012 Dec;31(12):1733-9. https://doi:10.1007/s10067-012-2080-7
- Punceviciene E, Gaizevska J, Sabaliauskaite R, Venceviciene L, Puriene A, Vitkus D, Jarmalaite S, Butrimiene I. Vitamin D and VDR Gene Polymorphisms’ Association with Rheumatoid Arthritis in Lithuanian Population. Medicina (Kaunas). 2021 Apr 3;57(4):346. https://doi:10.3390/medicina57040346
- Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, Birnbaum NS, Burmester GR, Bykerk VP, Cohen MD, Combe B, Costenbader KH, Dougados M, Emery P, Ferraccioli G, Hazes JM, Hobbs K, Huizinga TW, Kavanaugh A, Kay J, Kvien TK, Laing T, Mease P, Ménard HA, Moreland LW, Naden RL, Pincus T, Smolen JS, Stanislawska-Biernat E, Symmons D, Tak PP, Upchurch KS, Vencovsky J, Wolfe F, Hawker G. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 2010 Sep;69(9):1580-8. https://doi:10.1136/ard.2010.138461
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988 Mar;31(3):315-24. https://doi:10.1002/art.1780310302
- Prevoo ML, van ‘t Hof MA, Kuper HH, van Leeuwen MA, van de Putte LB, van Riel PL. Modified disease activity scores that include twenty-eight-joint counts. Development and validation in a prospective longitudinal study of patients with rheumatoid arthritis. Arthritis Rheum. 1995 Jan;38(1):44-8. https://doi:10.1002/art.1780380107
- Holick MF, Binkley NC, Bischoff-Ferrari HA, Gordon CM, Hanley DA, Heaney RP, Murad MH, Weaver CM; Endocrine Society. Evaluation, treatment, and prevention of vitamin D deficiency: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2011 Jul;96(7):1911-30. https://doi:10.1210/jc.2011-0385
- Metsalu T, Vilo J. ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015 Jul 1;43(W1):W566-70. https://doi.org/10.1093/nar/gkv468
- Cribbs A, Feldmann M, Oppermann U. Towards an understanding of the role of DNA methylation in rheumatoid arthritis: therapeutic and diagnostic implications. Ther Adv Musculoskelet Dis. 2015 Oct;7(5):206-19. https://doi:10.1177/1759720X15598307
- Bragazzi NL, Watad A, Neumann SG, Simon M, Brown SB, Abu Much A, Harari A, Tiosano S, Amital H, Shoenfeld Y. Vitamin D and rheumatoid arthritis: an ongoing mystery. CurrOpinRheumatol. 2017 Jul;29(4):378-388. https://doi:10.1097/BOR.0000000000000397
- Shirvani SS, Nouri M, Sakhinia E, Babaloo Z, Jadideslam G, Shahriar A, Farhadi J, Khabbazi A. The expression and methylation status of vitamin D receptor gene in Behcet’s disease. ImmunInflamm Dis. 2019 Dec;7(4):308-317. https://doi:10.1002/iid3.275
- Beckett E L, Duesing K, Martin C, Jones P, Furst J, King K, Lucock M. Relationship between methylation status of vitamin D-related genes, vitamin D levels, and methyl-donor biochemistry. J NutrIntermedMetab. 2016 (6): 8-15. https://doi.org/10.1016/j.jnim.2016.04.010
- Zhu H, Wang X, Shi H, Su S, Harshfield GA, Gutin B, Snieder H, Dong Y. A genome-wide methylation study of severe vitamin D deficiency in African American adolescents. J Pediatr. 2013 May;162(5):1004-9.e1. https://doi:10.1016/j.jpeds.2012.10.059
- Zhou Y, Zhao LJ, Xu X, Ye A, Travers-Gustafson D, Zhou B, Wang HW, Zhang W, Lee Hamm L, Deng HW, Recker RR, Lappe JM. DNA methylation levels of CYP2R1 and CYP24A1 predict vitamin D response variation. J Steroid Biochem Mol Biol. 2014 Oct;144 Pt A:207-14. https://doi:10.1016/j.jsbmb.2013.10.004
- Liu CC, Fang TJ, Ou TT, Wu CC, Li RN, Lin YC, Lin CH, Tsai WC, Liu HW, Yen JH. Global DNA methylation, DNMT1, and MBD2 in patients with rheumatoid arthritis. Immunol Lett. 2011 Mar 30;135(1-2):96-9. https://doi:10.1016/j.imlet.2010.10.003
- Karouzakis E, Gay RE, Michel BA, Gay S, Neidhart M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2009 Dec;60(12):3613-22. https://doi:10.1002/art.25018
Supplementary Material
Table S1. VDR, CYP24A1 and CYP2R1genes PCR and pyrosequencing primers
|
Primers sequences |
Length, nt |
%GC |
Amplicon length, bp |
|
|
VDR-s1 |
GTTGGGTTGTTTTTGTTTGTTAAAAG |
26 |
30.8 |
131 |
|
VDR-as2 |
Biotin-CCTATCCTAAAACCCCCTTTC |
21 |
47.6 |
|
|
VDR-seq3 |
TTGTTTTTGTTTGTTAAAAGG |
21 |
23.8 |
- |
|
CYP24A1-s1 |
AGTGTAAGGAGGTATTAATGTTTTGA |
26 |
30.8 |
130 |
|
CYP24A-as2 |
Biotin-AAAAAAACAAAAAAAACCAACTAATATAAT |
30 |
13.3 |
|
|
CYP24A1-seq3 |
GGAGGTATTAATGTTTTGAG |
20 |
35.0 |
- |
|
CYP2R1-s1 |
GGAAGTTTTGGAGAGTTGAAGAG |
23 |
43.5 |
222 |
|
CYP2R1-as2 |
Biotin-CCTCTCCCTACACCTAACTCTACTTTCT |
28 |
46.4 |
|
|
CYP2R1-seq3 |
AGTTGTTGAAGTAGAGG |
17 |
41.2 |
- |
PCR: polymerase chain reaction; nt: nucleotides; bp: base pairs; s1: sense primer; as2: antisense primer; seq3: pyrosequencing primer
Table S2. PCR cycling conditions
|
Temperature, ºC |
Duration |
Cycles |
Stage |
|
95 ºC |
15 min |
1 |
Enzyme activation |
|
95 ºC |
30 s |
45 |
DNA strand separation |
|
According to gene1 |
30 s |
Primer annealing |
|
|
72 ºC |
30 s |
DNA strand extension |
|
|
72 ºC |
10 min |
1 |
Synthesis completion |
|
4 ºC |
∞ |
1 |
Stopping/storing the reaction |
PCR, polymerase chain reaction;1 VDR – 58 ºC; CYP24A1 – 50 ºC;CYP2R1– 55 ºC